Intro
In this post, we will dive deeper into the {purrr} package. We will explore purrr::nest() %>% dplyr::mutate(), which is an alternative to the split() function in the base-R. Moreover, we will see how the combination of purrr::nest() %>% dplyr::mutate() with purrr::map2_*() or purrr::pmap_*() can be a powerful tool in functional programming.
The data we are going to use was narrowed down to the following variables of interest for the current post. For more information about data, please checkout here.
| Variable Code | Type | Description |
|---|---|---|
year |
String | The year that data was collected |
id |
Integer | Respondent ID |
age |
Integer | Age in years at screening |
gender |
Category | Gender: male or female |
race_ethnic |
Category | Race/Hispanic origin: mexican-american, non-hispanic-black, non-hispanic-white, other-hispanic, other-race |
hh_food_secure |
Integer | Household food security category over last 12 months |
age_group |
Binary | Age group: adult(20+) or child(6-19) |
educ |
Integer | Education level for adults 20+ and children/youth 6-19 |
Let’s take a look at the dataframe.
head(df)
# A tibble: 6 x 8
year id age gender race_ethnic hh_food_secure
<chr> <dbl> <dbl> <fct> <fct> <dbl>
1 1999-2000 2 77 male non-hispanic-white 1
2 1999-2000 3 10 female non-hispanic-white 1
3 1999-2000 5 49 male non-hispanic-white 1
4 1999-2000 6 19 female other-race 1
5 1999-2000 8 13 male non-hispanic-white 1
6 1999-2000 9 11 female non-hispanic-black 2
# … with 2 more variables: age_group <fct>, educ <dbl>split vs. nest
We are interested in how food security changes over time, separated by gender and age group (child and adult). For the first step, we want to split the dataframe into lists with split as the following. However, the three grouping variables (gender, age group, and year) are merged together into one column. This is less desirable because we will need each of these variables for later.
split_df = split(df, list(df$gender, df$age_group, df$year))
head(split_df$`female.adult.1999-2000`)
# A tibble: 6 x 8
year id age gender race_ethnic hh_food_secure
<chr> <dbl> <dbl> <fct> <fct> <dbl>
1 1999-2000 15 38 female non-hispanic-white 1
2 1999-2000 16 85 female non-hispanic-black 4
3 1999-2000 20 23 female mexican-american 1
4 1999-2000 24 53 female non-hispanic-white 1
5 1999-2000 25 42 female non-hispanic-white 1
6 1999-2000 34 38 female non-hispanic-black 3
# … with 2 more variables: age_group <fct>, educ <dbl>Equivalently, we can also use nest from the {purrr} package as the following:
nest_df = df %>%
group_by(year, age_group, gender) %>%
nest()
head(nest_df)
# A tibble: 6 x 4
# Groups: year, gender, age_group [6]
year gender age_group data
<chr> <fct> <fct> <list>
1 1999-2000 male adult <tibble [2,211 × 5]>
2 1999-2000 female child <tibble [1,698 × 5]>
3 1999-2000 male child <tibble [1,744 × 5]>
4 1999-2000 female adult <tibble [2,534 × 5]>
5 2001-2002 male adult <tibble [2,384 × 5]>
6 2001-2002 female adult <tibble [2,669 × 5]>These two approaches end with very similar results, except gender, age_group, and year are maintained with its original structure with nest() but not with split().
Moreover, with nest(), we can manipulate the data frame within each row while save the output as another column. We will calculate within each gender, age group and year, how food security changes with age. (If you find the map() function confusing, I encourage you take a look at this post.)
model_df = nest_df %>%
mutate(n = map_dbl(data, nrow),
m1 = map(data, ~lm(hh_food_secure ~ age, data = .x)),
coefs = map(m1, coef),
intercept = map_dbl(coefs, 1),
slope = map_dbl(coefs, 2))
head(model_df)
# A tibble: 6 x 9
# Groups: year, gender, age_group [6]
year gender age_group data n m1 coefs intercept
<chr> <fct> <fct> <list> <dbl> <list> <list> <dbl>
1 1999-2000 male adult <tibble [2… 2211 <lm> <dbl … 1.599724
2 1999-2000 female child <tibble [1… 1698 <lm> <dbl … 1.573725
3 1999-2000 male child <tibble [1… 1744 <lm> <dbl … 1.691323
4 1999-2000 female adult <tibble [2… 2534 <lm> <dbl … 1.582938
5 2001-2002 male adult <tibble [2… 2384 <lm> <dbl … 1.752420
6 2001-2002 female adult <tibble [2… 2669 <lm> <dbl … 1.703107
# … with 1 more variable: slope <dbl>first figure
Let’s take a look at how the slope change with time:
model_df %>%
ggplot(aes(x = year, y = slope, color = age_group, group = age_group)) +
geom_line(size = 1.5) +
facet_wrap(~gender, nrow = 2) +
theme(legend.position = 'bottom') +
labs(y = 'Slope: Age and Food Security',
x = 'Year',
color = 'Age Group')

From the figure, we can see that for adults, the slope is consistently negative across all time. In other words, as people age, the food security score decreased. However, for children, the food security increased dramatically recently, especially since 2010s. What if we want to dig deeper and see how age influences children’s food security within each year for different gender?
Let’s start with only one row of data:
child_model_df = model_df %>% filter(age_group == 'child')
plotting <- function(df, gender, year = NULL){
p = df %>%
group_by(age, race_ethnic) %>%
summarise(m = mean(hh_food_secure),
sd = sd(hh_food_secure)) %>%
ggplot(aes(x=age,
y=m,
color = race_ethnic))+
geom_line(alpha = 0.7, size = 1.5) +
theme(legend.position = 'bottom')+
labs(x = 'Age',
y = 'Food Secure Score',
color = 'Race and Ethnic') +
guides(color=guide_legend(nrow=2, byrow=TRUE))
if(missing(year)){
p = p + labs(title = gender)
}else{
p = p + labs(title = paste(year, gender, sep = ": "))}
p
}
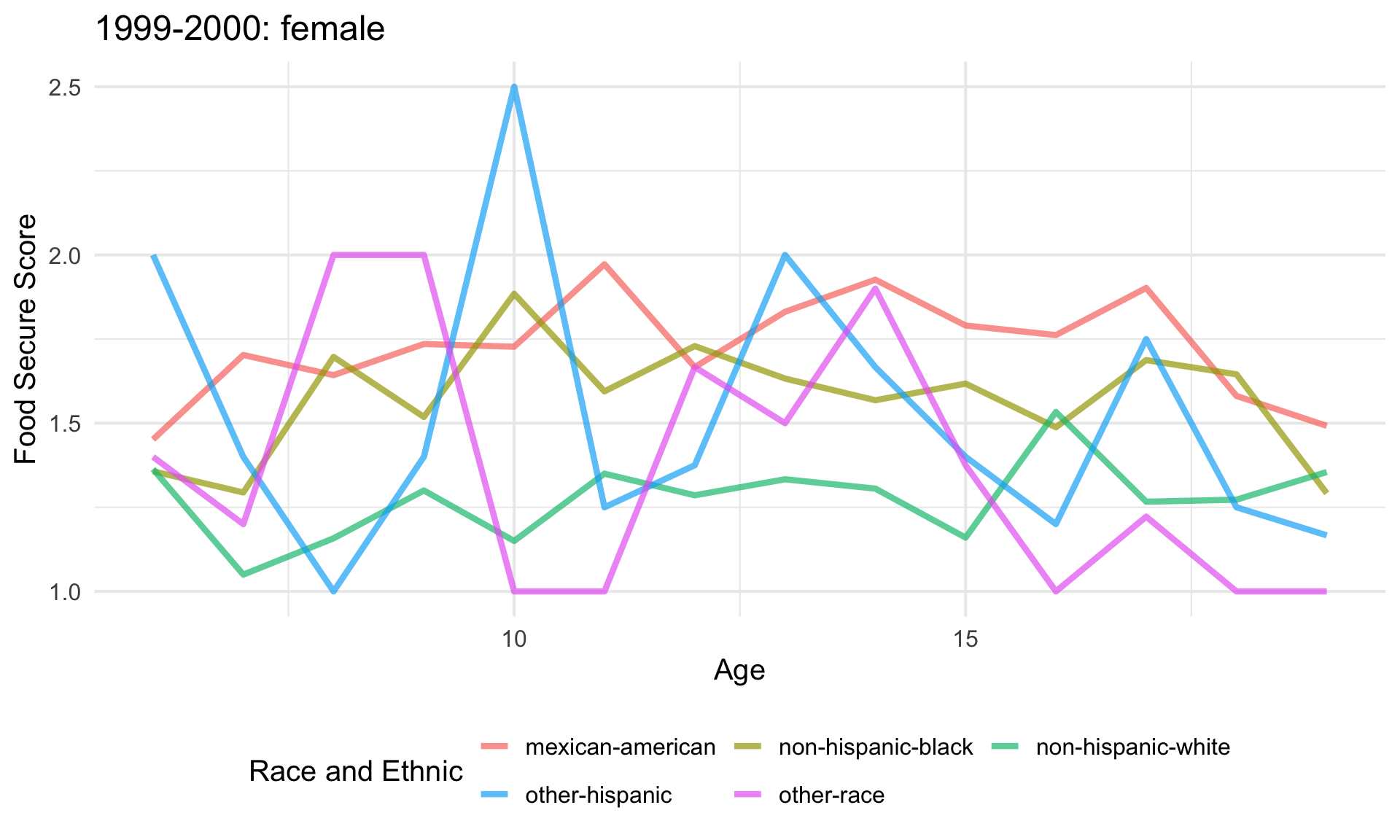
# make sure the funtion works for one row of data.
plotting(child_model_df$data[[1]],
child_model_df$gender[[1]],
child_model_df$year[[1]])
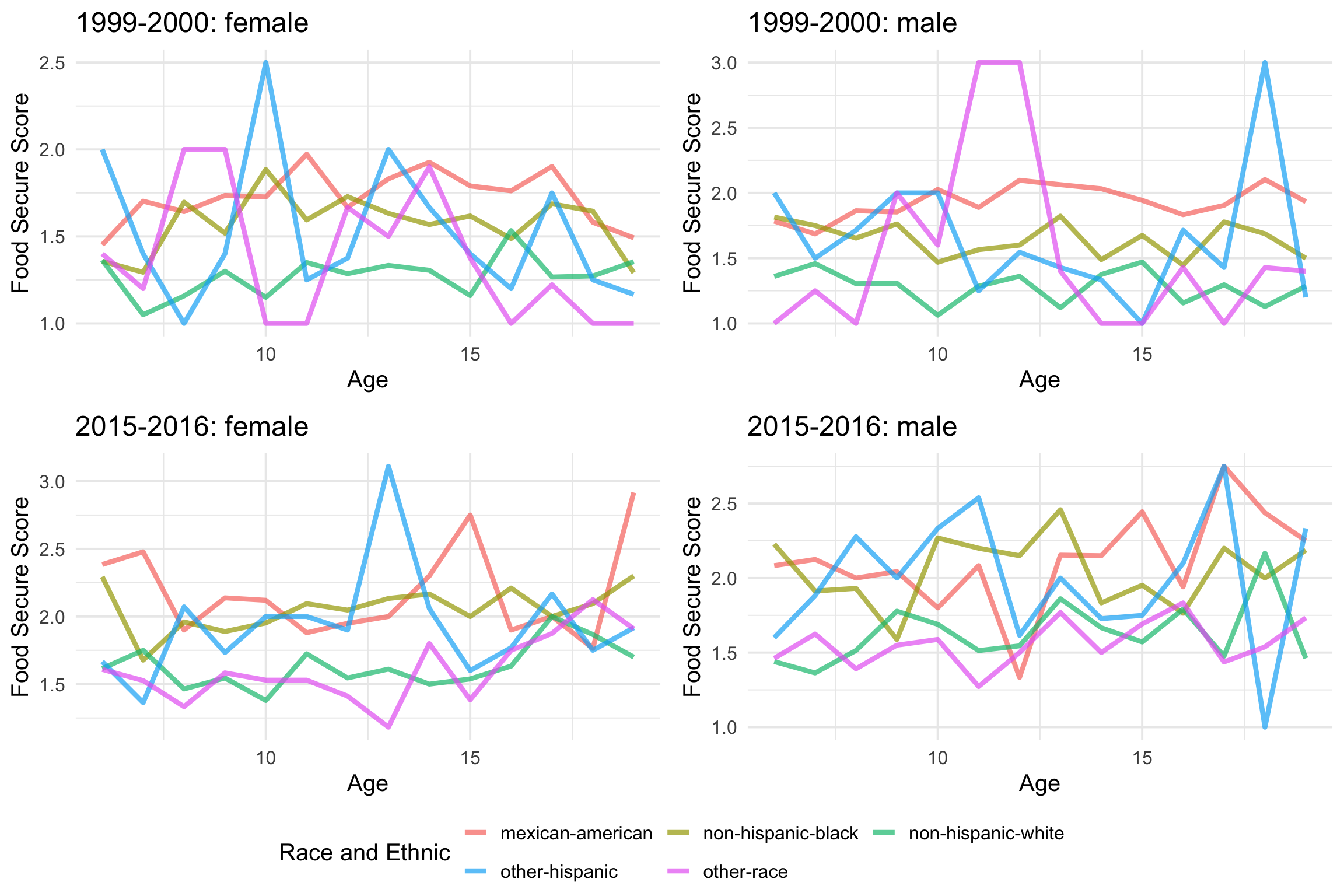
pmap_*
With pmap_*, we can easily use the above code to produce figures for each row. When using pmap_*, the first input is a list of column names that we need from the dataframe and the second input is the plotting function we used in the last part. The ..1, ..2, and ..3 are corresponding to data, gender, and year. Then, Voila, you have a figure for data from each row!
child_model_plot_df <- child_model_df %>%
mutate(nest_plot = pmap(list(data, gender, year),
~{plotting(..1, ..2, ..3)})
)
ggpubr::ggarrange(child_model_plot_df$nest_plot[[1]],
child_model_plot_df$nest_plot[[2]],
child_model_plot_df$nest_plot[[17]],
child_model_plot_df$nest_plot[[18]],
ncol = 2, nrow = 2,
common.legend = TRUE,
legend = 'bottom')
Another cool thing about nest, is that we can easily reverse this process with unnest after we finished our grouped analysis.
adult_df = model_df %>%
filter(age_group == 'adult') %>%
select(year, gender, age_group, data) %>% unnest(data)
head(adult_df)
# A tibble: 6 x 8
# Groups: year, gender, age_group [1]
year gender age_group id age race_ethnic
<chr> <fct> <fct> <dbl> <dbl> <fct>
1 1999-2000 male adult 2 77 non-hispanic-white
2 1999-2000 male adult 5 49 non-hispanic-white
3 1999-2000 male adult 12 37 non-hispanic-white
4 1999-2000 male adult 13 70 mexican-american
5 1999-2000 male adult 14 81 non-hispanic-white
6 1999-2000 male adult 29 62 non-hispanic-white
# … with 2 more variables: hh_food_secure <dbl>, educ <dbl>map2_*
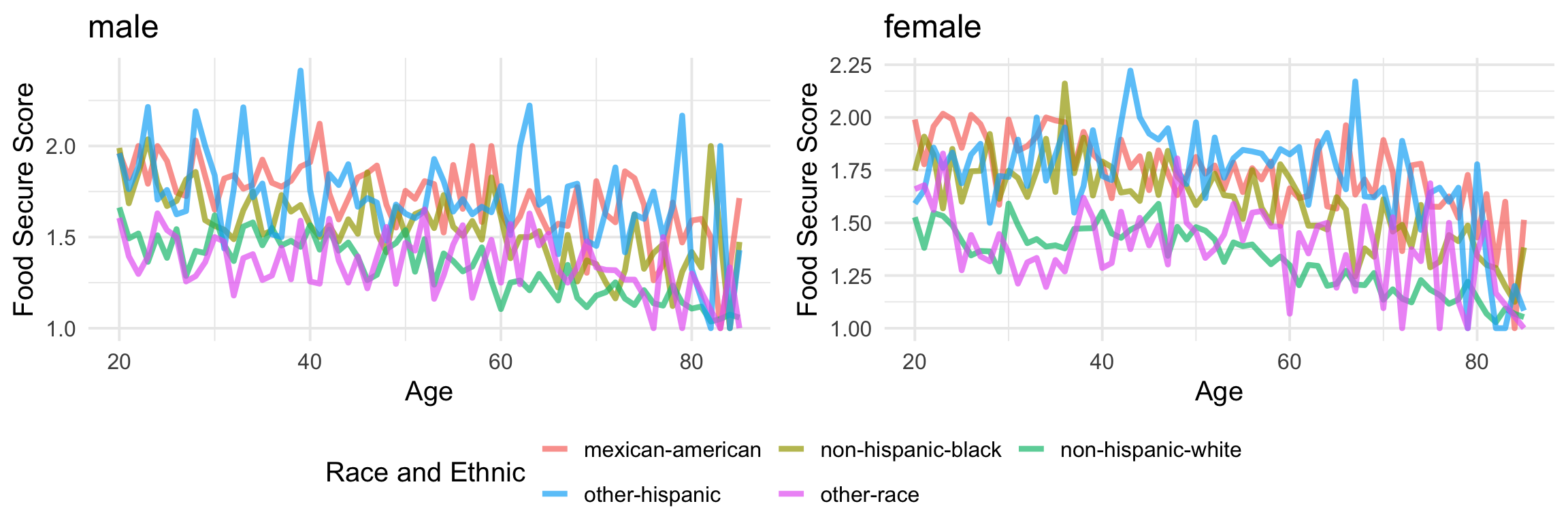
Lastly, once we learned about pmap_*, map2_* is very similar. Instead of being able to use as many variables you need with pmap, map2 is specialized for only 2 inputs. Let’s see an example below. Since we have found that the relationship between age and food security are pretty consistent over the years, let’s make a plot that ignore the age factor with map2.
adult_plot_df = adult_df %>%
group_by(gender) %>%
nest() %>%
mutate(nest_plot = map2(data, gender,
~plotting(.x, .y)))
ggpubr::ggarrange(adult_plot_df$nest_plot[[1]],
adult_plot_df$nest_plot[[2]],
ncol = 2,
common.legend = TRUE,
legend = 'bottom')